How genetic motifs conduct "the music of life"
Using AI and supercomputers, researchers have discovered reoccurring patterns and combinations, known as ‘motifs’, of the four molecular building blocks A, C, G and T, connecting them to gene expression, that is, average amounts of produced proteins.
Our genetic codes control not only which proteins our cells produce, but also - to a great extent - in what quantity. This ground-breaking discovery, applicable to all biological life, was recently made by systems biologists at Chalmers University of Technology, Sweden, usingsupercomputersandartificial intelligence. Their research could shed new light on the mysteries of cancer.
DNA molecules contain instructions for cells for producing various proteins. This has been known since the middle of the last century when the double helix was identified as the information carrier of life.
But until now, the factor which determines what quantity of a certain protein will be produced has been unclear. Measurements have shown that a single cell can contain anything from a few molecules of a given protein, up to tens of thousands.
With this new research, our understanding of the mechanisms behind this process, known as gene expression, has taken a big step forward. The group of Chalmers scientists have shown that most of the information for quantity regulation is also embedded in the DNA code itself. They have demonstrated that this information can be read with the help of supercomputers and AI.
Comparable to an orchestral score
Assistant Professor Aleksej Zelezniak, of Chalmers' Department of Biology and Biological Engineering, leads the research group behind the discovery. "You could compare this to an orchestral score. The notes describe which pitches the different instruments should play. But the notes alone do not say much about how the music will sound," he explains.
Information for the tempo and dynamics of the music are also required, for example. But instead of written instructions such as allegro or forte in connection with the notation, the language of genetics spreads this information over large areas of the DNA molecule. "Previously, we could read the notes, but not how the music should be played. Now we can do both," states Aleksej Zelezniak. "Another comparison could be that now we have found the grammar rules for the genetic language, where perhaps before we only knew the vocabulary."
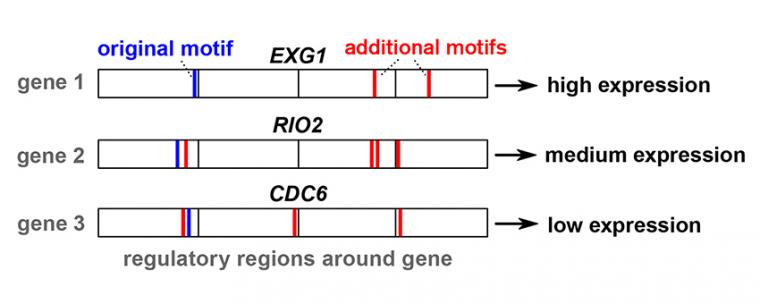
What then is this grammar, which determines the quantity of gene expression? According to Aleksej Zelezniak, it takes the form of reoccurring patterns and combinations of the four 'notes' of genetics - the molecular building blocks designated A, C, G and T. These patterns and combinations are known as 'motifs'.
The crucial factors are the relationships between these motifs - how often they repeat and at exactly which positions in the DNA code they appear. "We discovered that this information is distributed over both the coding and non-coding parts of DNA - meaning, it is also present in the areas that used to be referred to as 'junk DNA'."
A discovery that applies to all biological life
Although there are other factors that also affect cells' gene expression, according to the Chalmers researchers' study, the information embedded in the genetic code accounts for about 80 per cent of the process.
The researchers tested the method in seven different model organisms - from yeast andbacteriato fruit flies, mice, and humans - and found that the mechanism is the same. The discovery they have made is universal, valid for all biological life.
According to Aleksej Zelezniak, the discovery would have not been possible without access to state-of-the-art supercomputers and AI. The research group conducted huge computer simulations both at Chalmers University of Technology and other facilities in Sweden. "This tool allows us to look at thousands of positions at the same time, creating a kind of automated examination of DNA. This is essential for being able to identify patterns from such huge amounts of data."
Jan Zrimec, postdoctoral researcher in the Chalmers group and first author of the study, agrees, saying: "With previous technologies, researchers had to tell the system which motifs in the DNA code to search for. But thanks to AI, the system can now learn on its own, identifying different motifs and motif combinations relevant to gene expression."
He adds that the discovery is also due to the fact they were examining a much larger part of DNA in a single sweep than had previously been done.
Fast value for the pharmaceutical industry
Aleksej Zelezniak believes that the discovery will generate great interest in the research world, and that the method could become an important tool in several research fields - genetics and evolutionary research, systems biology, medicine, and biotechnology.
The new knowledge could also make it possible to better understand how mutations can affect gene expression in the cell and therefore, eventually, how cancers arise and function. The applications which could most rapidly be significant for the wider public are in the pharmaceutical industry.
"It is conceivable that this method could help improve the genetic modification of themicroorganisms今天已经使用“生物工厂”——李ding to faster and cheaper development and production of new drugs," he speculates.
The research was published inNature Communications.